DrugSimDB: A Comprehensive Resource for Drug Similarity and Repositioning
Published in Briefings in Bioinformatics (2021)
Read the paper here
Download full database
Why Drug Similarity Matters
Drug discovery is notoriously expensive, time-consuming, and risky. Traditional pipelines often take over a decade and billions of dollars to bring a new molecule to market.
An emerging solution is drug repositioning: finding new therapeutic uses for existing drugs. This approach bypasses several costly development stages and leverages already-known safety profiles.
At the heart of repositioning lies a simple but powerful idea:
Drugs with similar properties are likely to have similar therapeutic effects.
But how do we systematically measure similarity between drugs?
This is where DrugSimDB comes in.
Introducing DrugSimDB
DrugSimDB is a comprehensive, integrative drug–drug similarity database, covering over 10,317 small-molecule drugs (including 2,466 approved drugs) and providing more than 238,000 significant drug–drug similarity pairs.
Unlike existing resources that rely on one type of evidence (e.g., chemical structure or side effects), DrugSimDB integrates multiple modalities of evidence:
- Chemical similarity (molecular fingerprints, atom-pair Tanimoto scores)
- Target protein sequence similarity
- Target functional similarity (Gene Ontology enrichment + PPI context)
- Pathway-based similarity (shared KEGG pathways and constituent genes)
By aggregating these diverse signals, DrugSimDB delivers robust, multi-modal similarity scores that outperform single-source approaches in predicting repositioning candidates.
How It Works
Methodological Overview
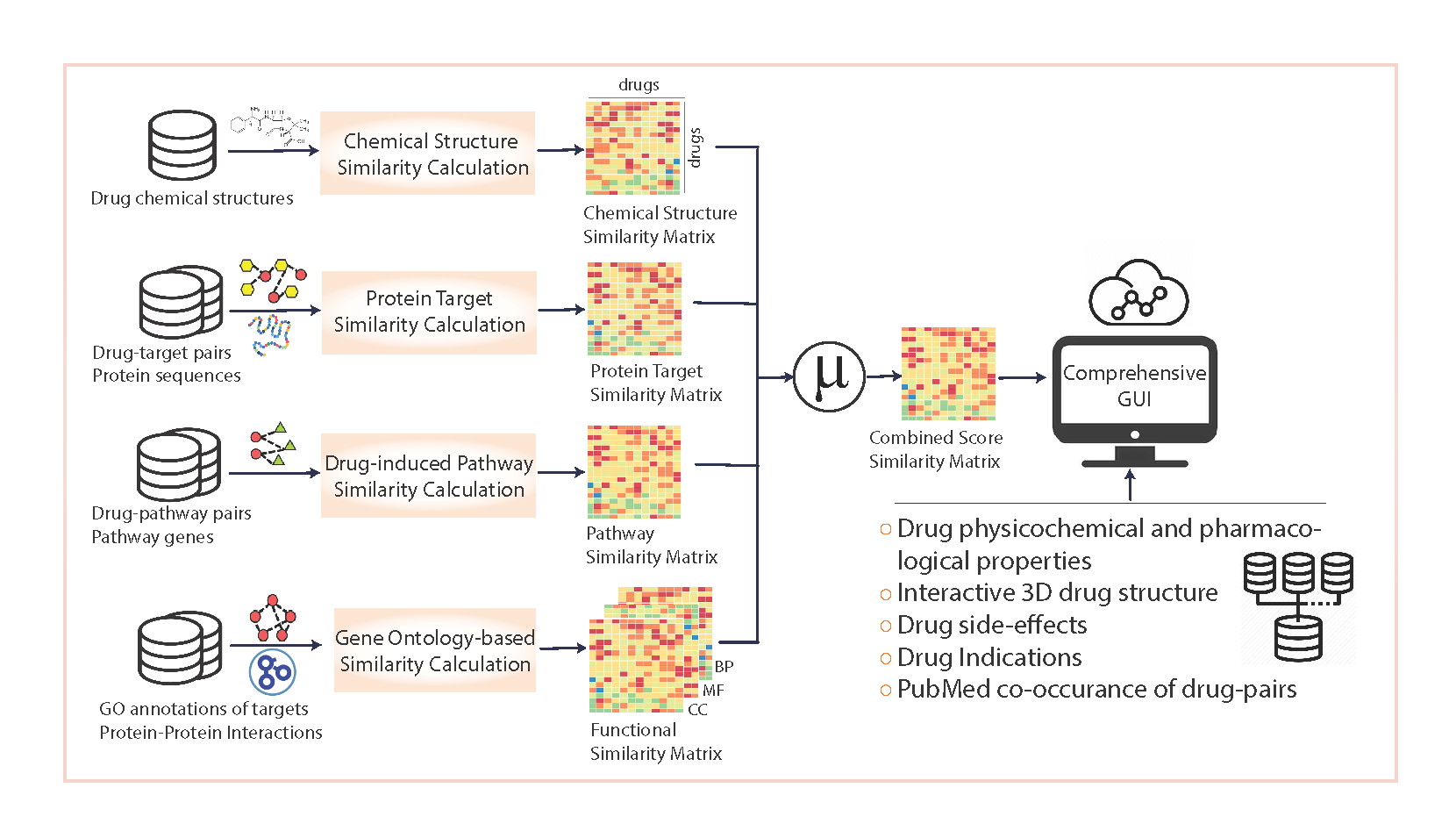
Figure: Multi-layer integration of drug similarity measures into a unified resource.
- Data collection – integrates DrugBank, KEGG, I2D (protein interactions), EnrichR, RepoDB, and SIDER.
- Computation – calculates drug similarity across multiple modalities in R (parallelized for efficiency).
- Integration – aggregates similarity matrices into a single multi-evidence drug similarity network.
- Access – provides a web interface and bulk-download options.
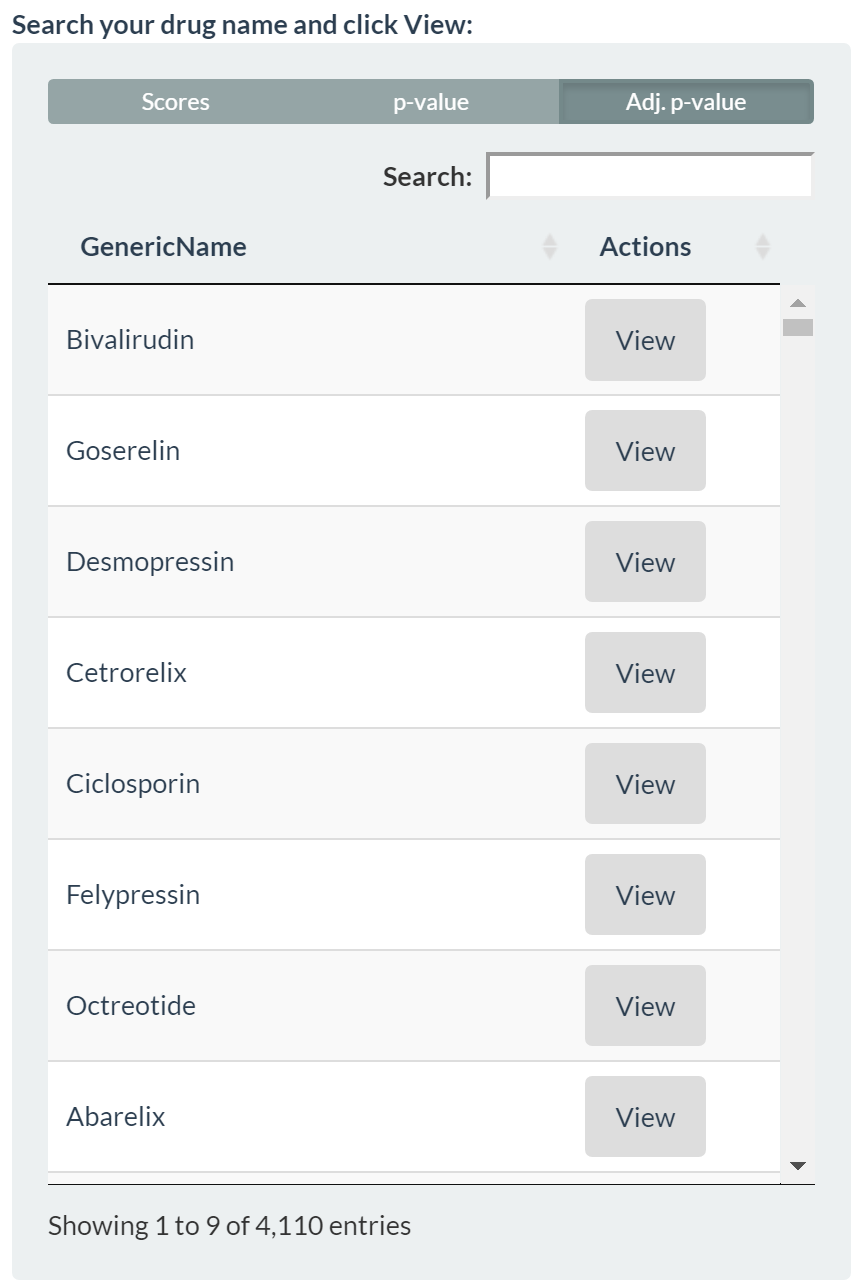
Interactive Web Server
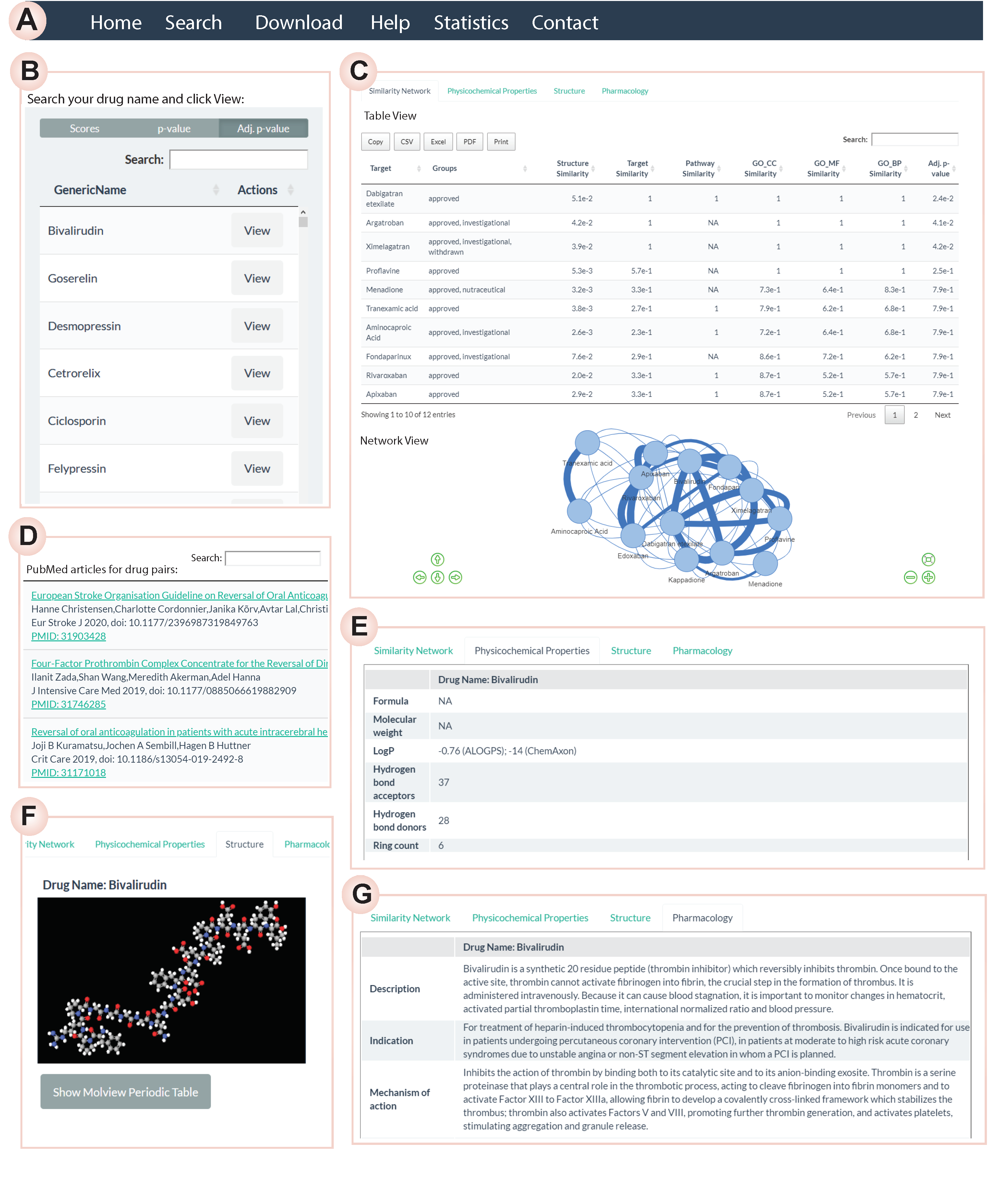
Figure: Web server The interactive web app (built with R Shiny) allows users to:
Search a drug and inspect its side-effects (from SIDER)
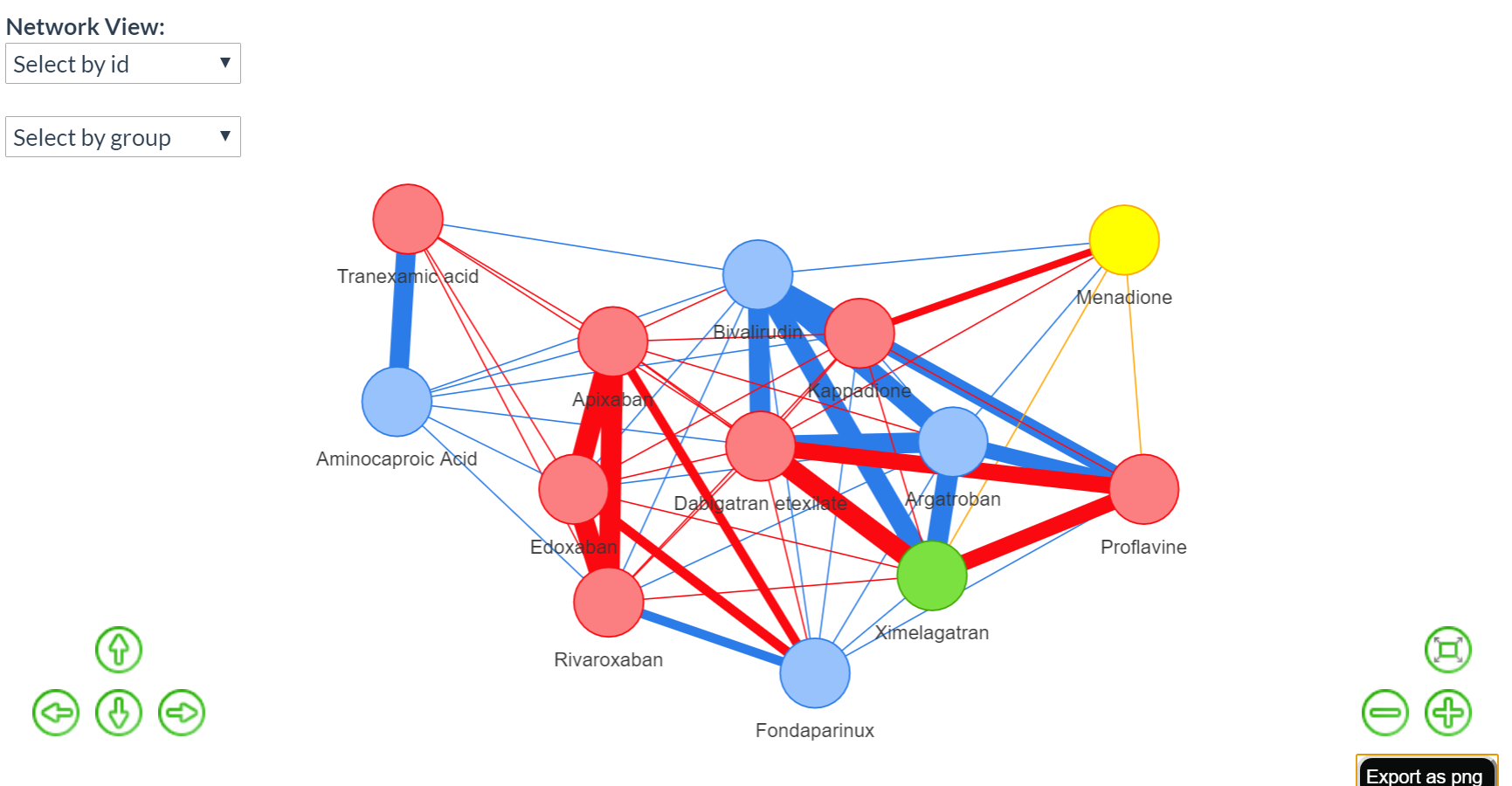
Explore its similarity network
See the tabular view
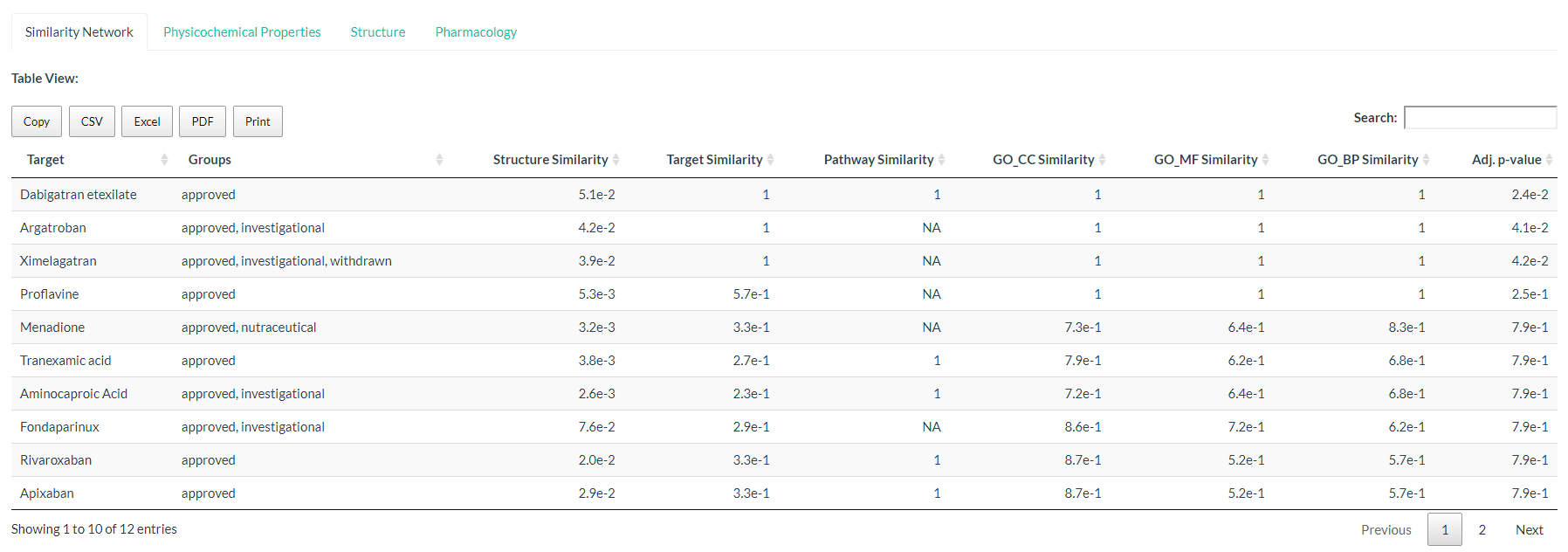 .
.Extract a drug information by just hovering your mouse on the drug node
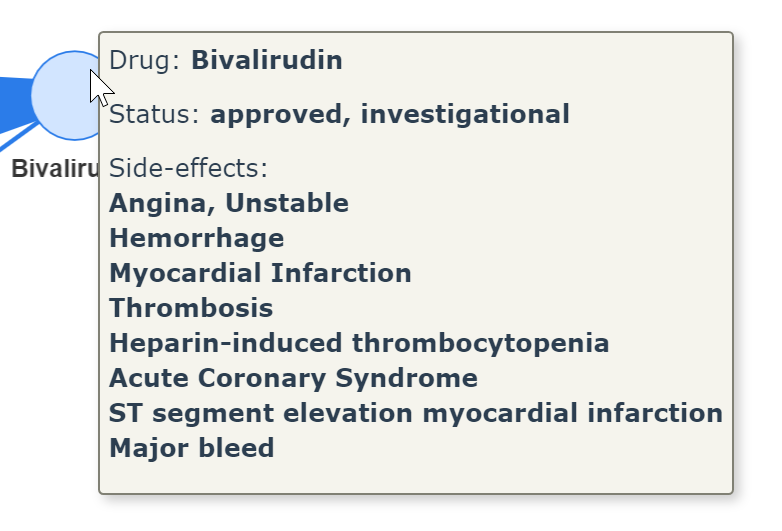 .
.Retrieve PubMed evidence for drug-pair associations by just clicking/tapping on an edge
 .
.Visualize 3D chemical structures
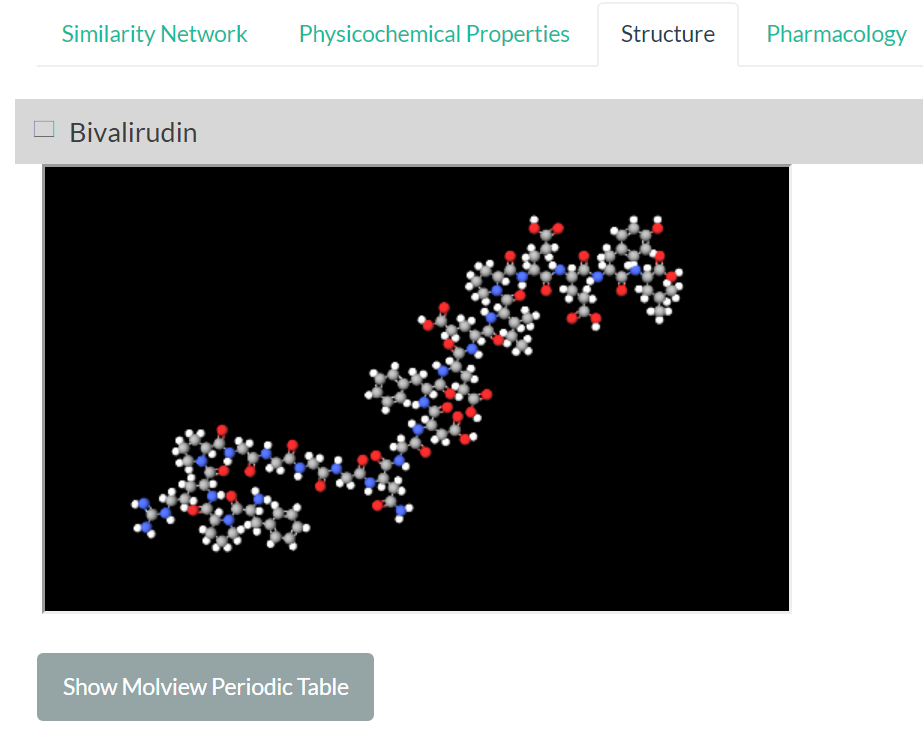 .
.Browse physiochemical & pharmacological properties
 .
.
What Makes DrugSimDB Novel?
DrugSimDB addresses three critical gaps in existing drug similarity resources:
- Coverage
- Over 10,000 small-molecule drugs, covering both approved and experimental compounds.
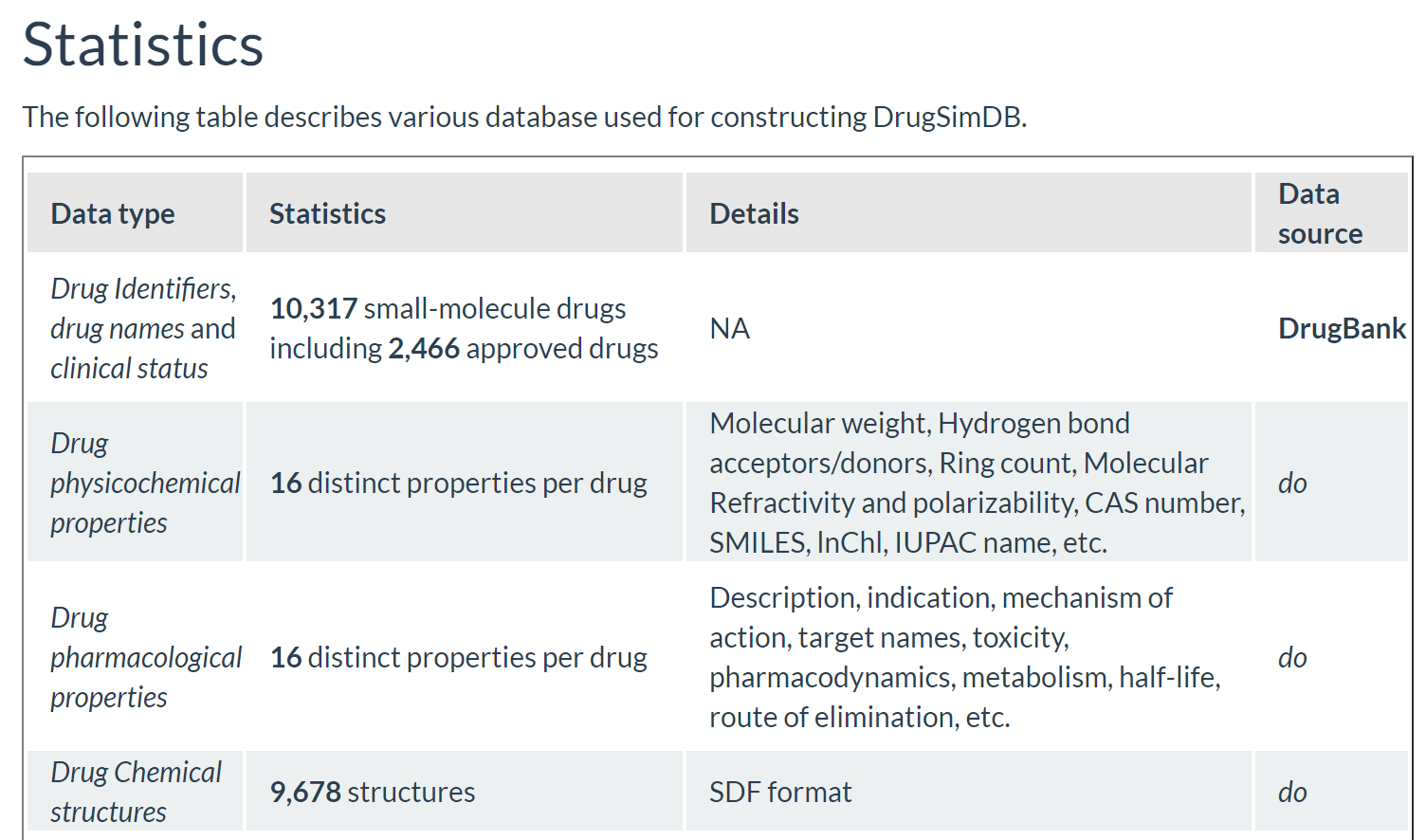
- Over 10,000 small-molecule drugs, covering both approved and experimental compounds.
- Integration
- Combines chemical, target, functional, and pathway evidence, ensuring better coverage of sparsely annotated drugs.
- Network-Based Validation
- The resulting similarity network follows a scale-free topology, like many biological networks.
- Validation against RepoDB repositioning gold standards yields a competitive AUC of 0.708, surpassing single-modality methods.
Key Results
- Scale-free topology: A few hub drugs link the network, enabling powerful diffusion-based repositioning methods.
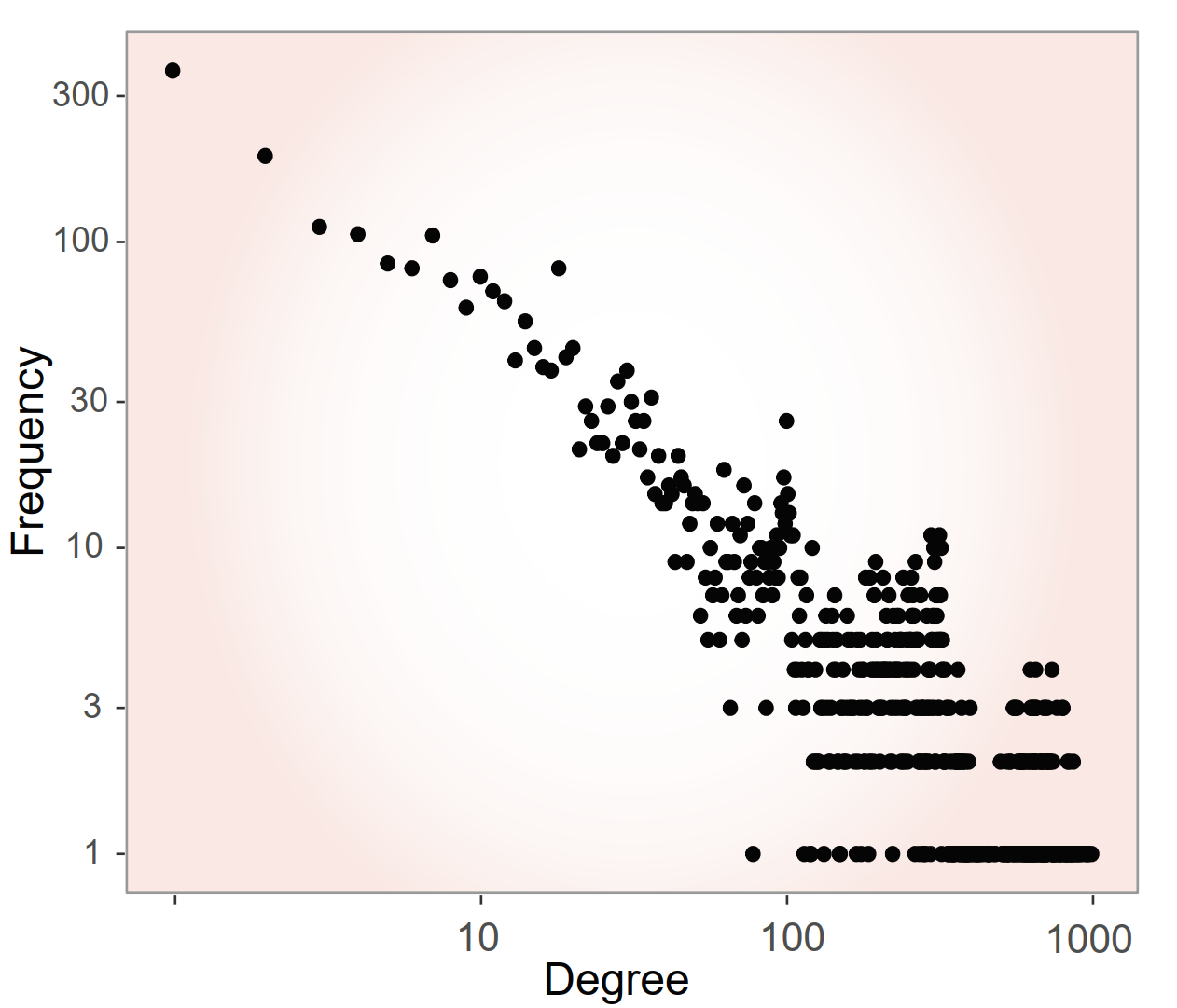
- Reduced data sparsity: Multi-source integration fills gaps where single data types are missing.
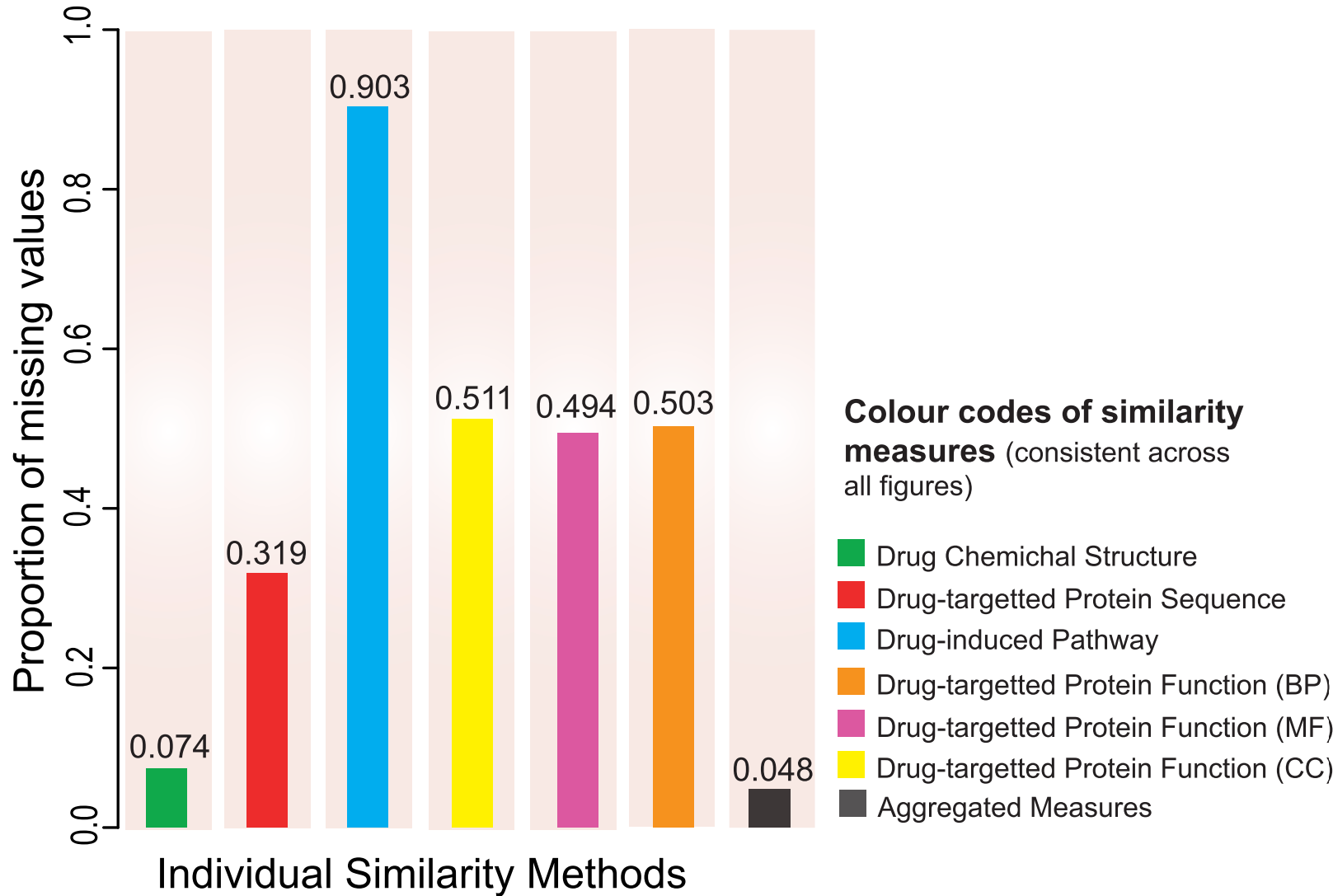
- Improved repositioning performance: Multi-modal scores outperform individual similarity metrics.
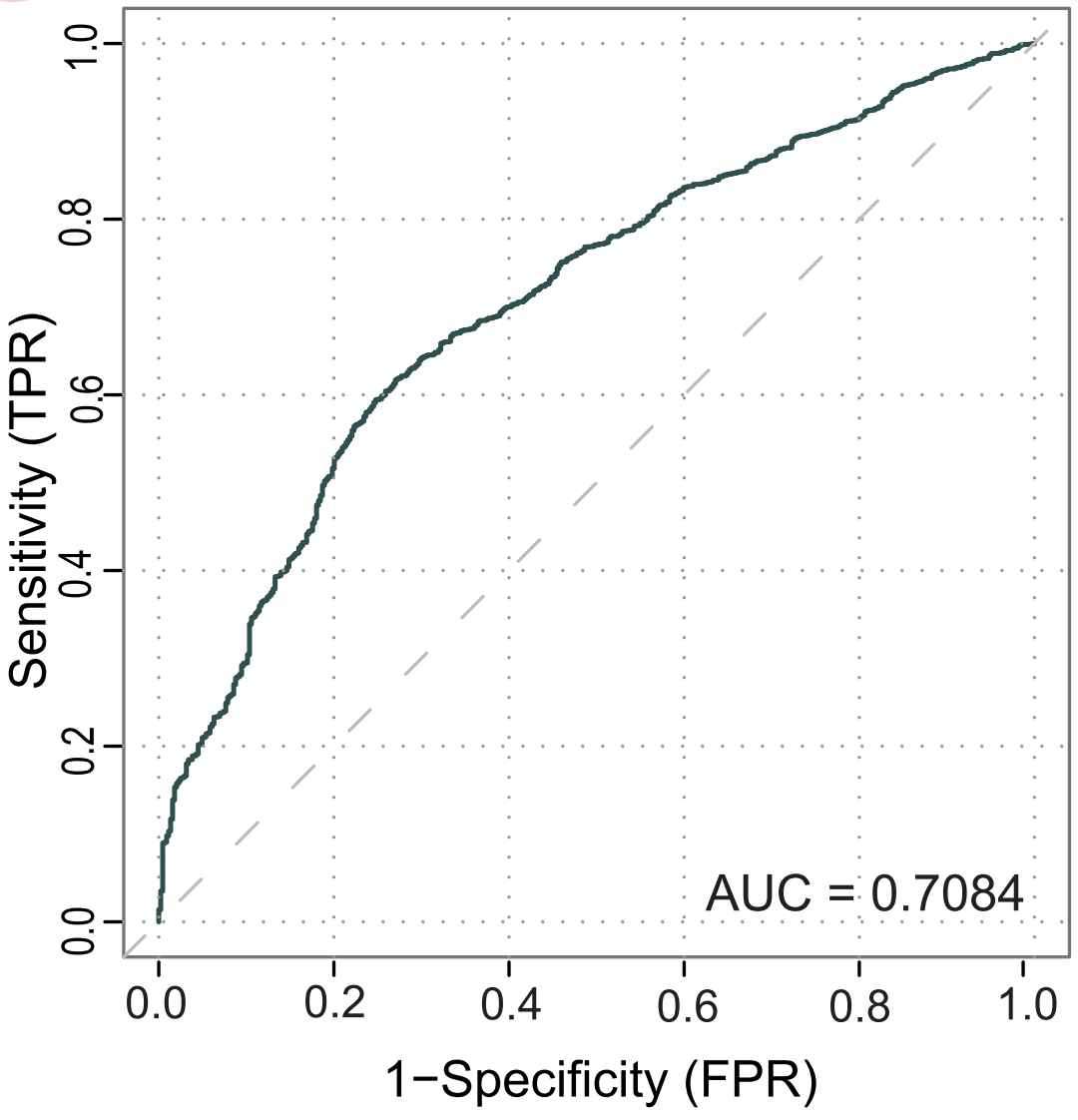 . Also the ROC plot for the further validation.
. Also the ROC plot for the further validation. 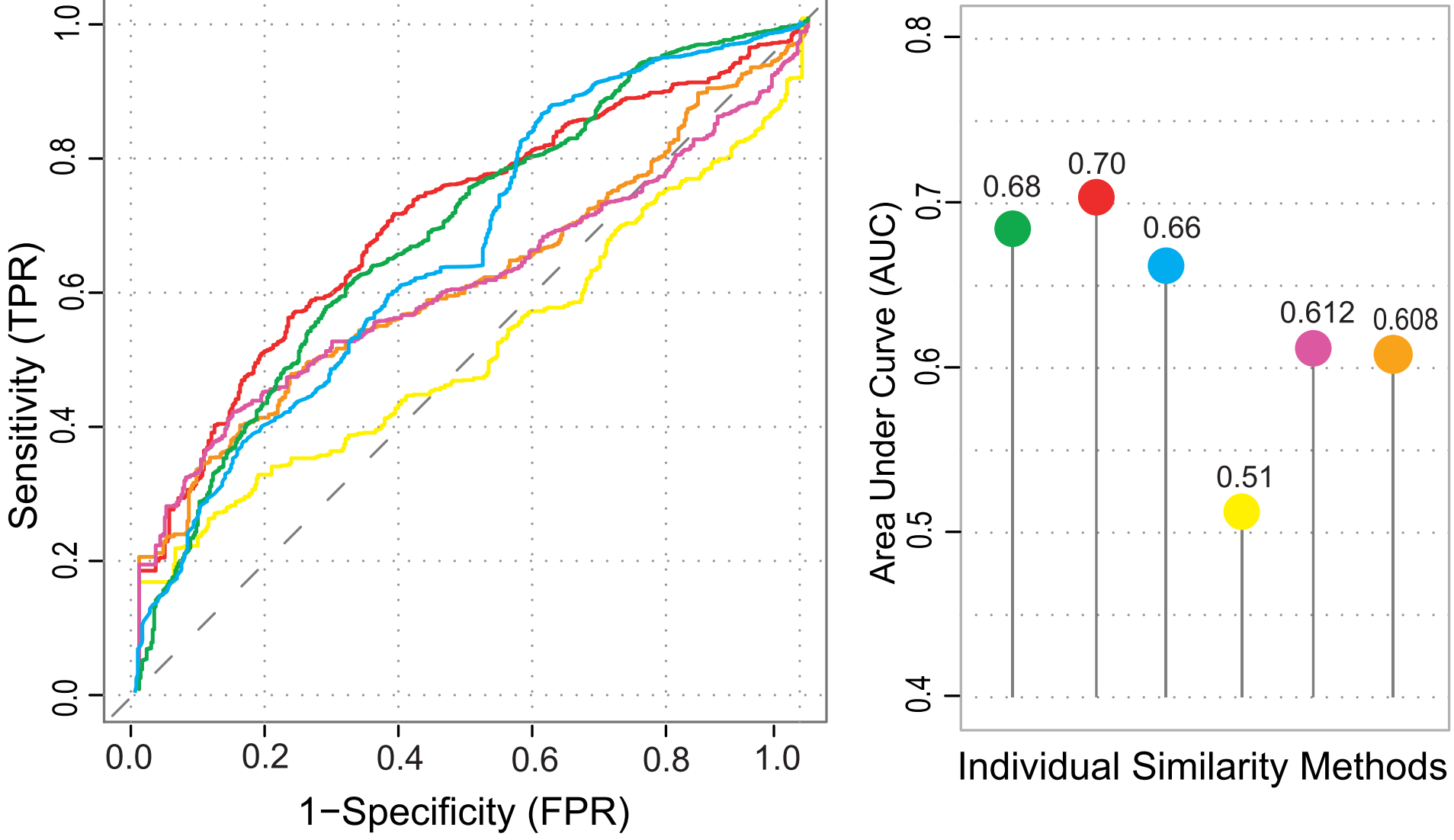
Figure: ROC analysis shows DrugSimDB’s integrated similarity scores outperform single-source measures.
Usage Examples
DrugSimDB is very easy to use. 
- Drug repositioning
- Identify candidate drugs for new indications by querying similarity to known therapeutics.
- Side-effect prediction
- Explore overlapping drug profiles to anticipate adverse events.
- Drug–drug interaction (DDI) analysis
- Investigate potential pharmacological conflicts in polypharmacy.
- Network medicine approaches
- Apply graph algorithms (community detection, random walks, diffusion) to uncover hidden drug relationships.
Future Potentials
DrugSimDB is designed to be flexible and extendable. Future updates may include:
- Integration of omics-driven signatures (transcriptomic, metabolomic).
- Incorporation of secondary/3D target structures.
- Deeper annotation with real-world evidence (EHRs, adverse-event reports).
- Network-based AI applications (e.g., graph neural networks for drug repositioning).
By serving as a foundational drug similarity knowledgebase, DrugSimDB can fuel discoveries in:
🧬 Rare Disease Therapeutics
Rare diseases often lack effective treatments because traditional drug discovery pipelines are not economically attractive for small patient populations. DrugSimDB offers a shortcut by enabling systematic searches for existing drugs with similar molecular profiles to those used in rare conditions. For example, if an experimental compound shows promise in a rare neurodegenerative disorder, DrugSimDB can help identify already-approved drugs with overlapping targets, pathways, or functional signatures. This could accelerate access to therapies for underserved patient groups while reducing development costs.
🎯 Cancer Precision Medicine
Cancer therapy increasingly relies on precision medicine, where treatments are tailored to the molecular characteristics of a patient’s tumor. DrugSimDB’s integration of target sequence, pathway, and functional similarities makes it a valuable tool for uncovering alternative therapeutic options when resistance develops. For instance, if a cancer drug fails due to acquired resistance, its similarity neighbors in DrugSimDB may reveal backup candidates that act on convergent pathways. This could guide oncologists in designing drug substitution or combination strategies based on molecular evidence.
🥗 Drug–Nutrient and Drug–Microbiome Interactions
The emerging fields of nutritional pharmacology and microbiome research highlight how diet and gut bacteria influence drug efficacy and safety. By extending DrugSimDB’s similarity framework, researchers could explore how drugs with shared metabolic or functional pathways might interact with nutrients or microbial metabolites. This opens doors to predicting nutrient–drug interactions (e.g., supplements that enhance or interfere with drug action) and drug–microbiome crosstalk (e.g., antibiotics altering microbiome-driven drug metabolism). Such insights could contribute to more holistic, personalized treatment plans.
🌐 Systems Pharmacology Models
Systems pharmacology seeks to understand drug effects at the network level, integrating multi-omics data, signaling pathways, and phenotypic responses. Because DrugSimDB itself is a scale-free drug similarity network, it naturally lends itself to graph-based modeling. Researchers can overlay gene expression, metabolomic, or clinical data onto DrugSimDB to perform network diffusion, community detection, and multi-layer modeling. This would support the development of computational pipelines that simulate how drugs perturb biological systems, guiding both mechanism-of-action studies and rational polypharmacy design.
Access and Citation
- 🌐 Explore the web app: DrugSimDB
- 📦 Download the source code & database: GitHub Repo
- 📖 Cite the paper:
Azad AKM, Dinarvand M, Nematollahi A, Swift J, Lutze-Mann L, Vafaee F.
A comprehensive integrated drug similarity resource for in-silico drug repositioning and beyond.
Briefings in Bioinformatics. 2021; 22(3): bbaa126. doi:10.1093/bib/bbaa126
Final Thoughts
DrugSimDB is more than a database—it’s a platform for innovation in drug discovery and repositioning. By lowering the barrier to similarity-based exploration, it empowers researchers worldwide to repurpose old drugs, predict new interactions, and accelerate precision medicine.
If you are working on drug discovery, computational pharmacology, or systems biology, DrugSimDB is a resource you don’t want to miss.